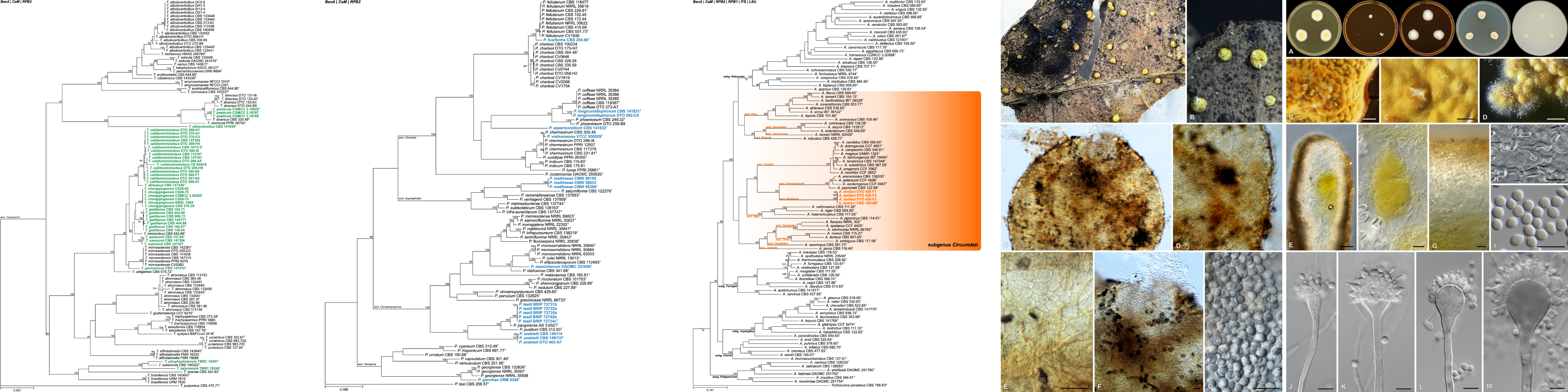
Aim: To understand the molecular mechanisms of plant resistance in the cereal pearl millet
 |
 |
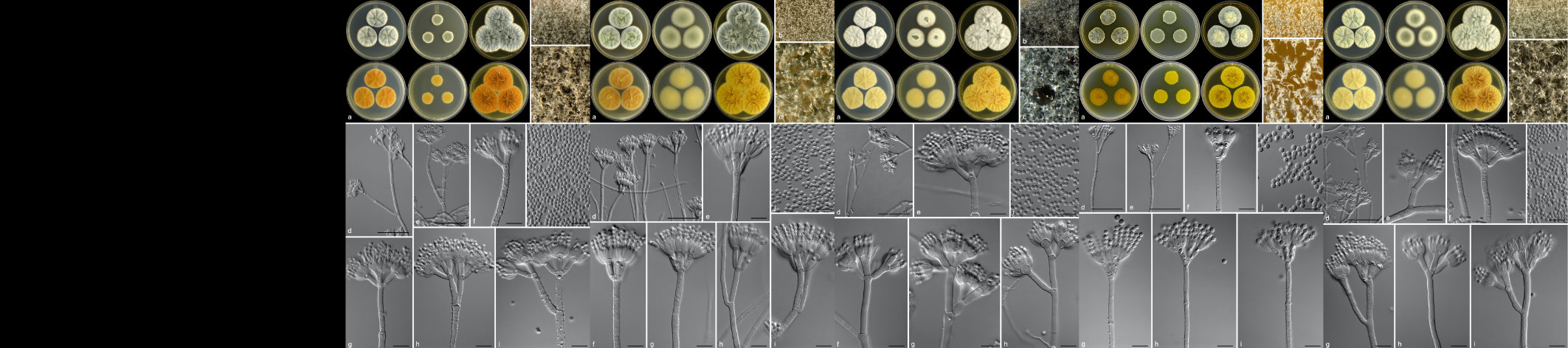
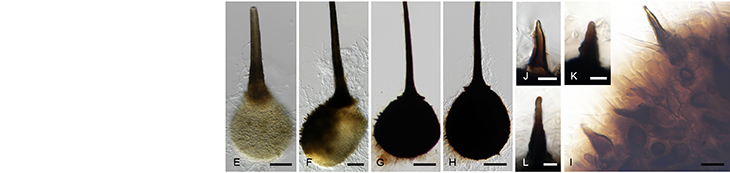
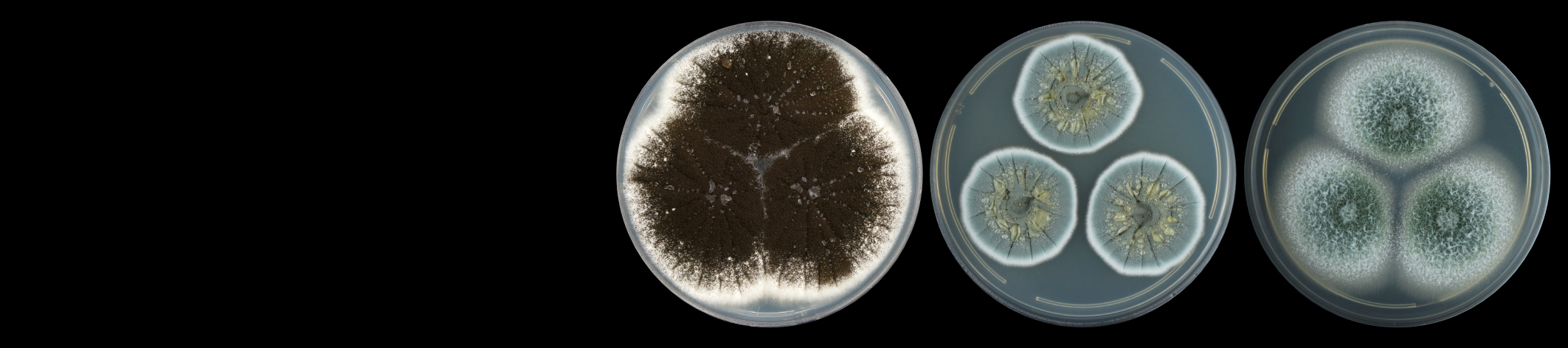
Pearl millet [Pennisetum glaucum (L.) R. Br.] is a cereal of importance to food security in Africa since the varieties planted are more tolerant to drought stress than other cereals. The main biotic stresses are downy mildew and rust diseases. The aim of the research project is to identify novel mechanisms of defense against stress through studies of the millet transcriptome. Since, very little genome sequence is available for this non-model crop, we chose to construct cDNA libraries from pearl millet using the Suppressive Subtraction Hybridization (SSH) technique. A novel method was developed for screening these SSH cDNA libraries on microarray slides (Van den Berg et al. 2004). This method allows identification of up-regulated genes, as well as determining whether the up-regulated transcripts are rare or abundant in the original treated sample. A complete methods protocol for library construction and description of a software package SSHscreen in R language have been published in a book chapter in 2007 (Berger et al. 2007).
Gene expression profiling of pearl millet treated with a range of defence signaling molecules as well as pathogen infection has been completed by Bridget Crampton at CSIR-Bio/Chemtek, and candidate defence genes are under investigation.

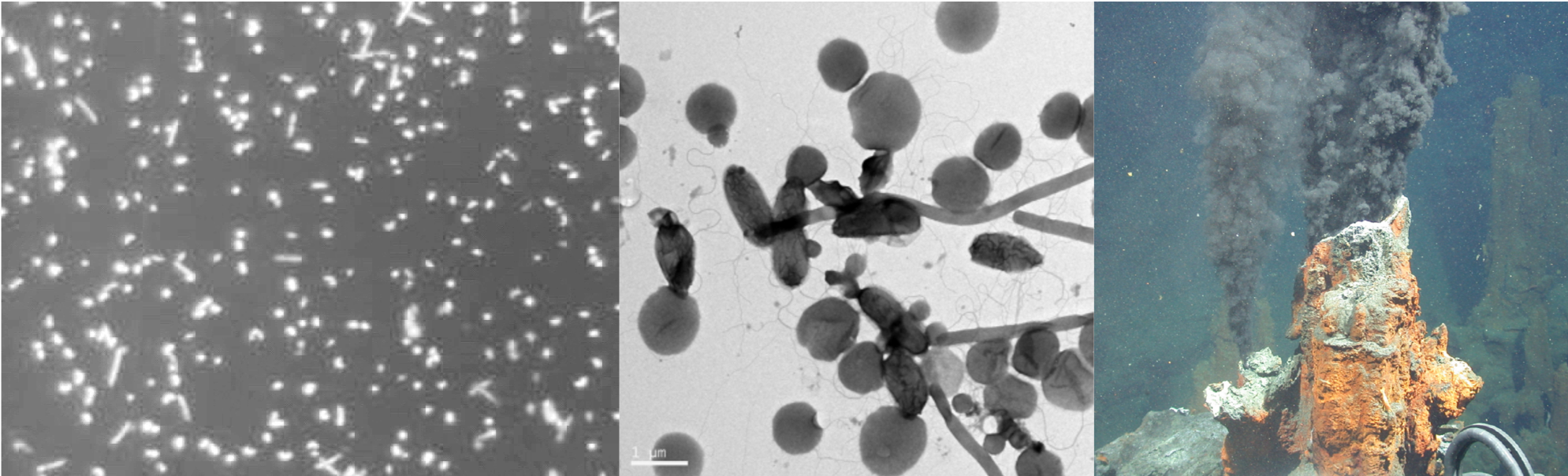
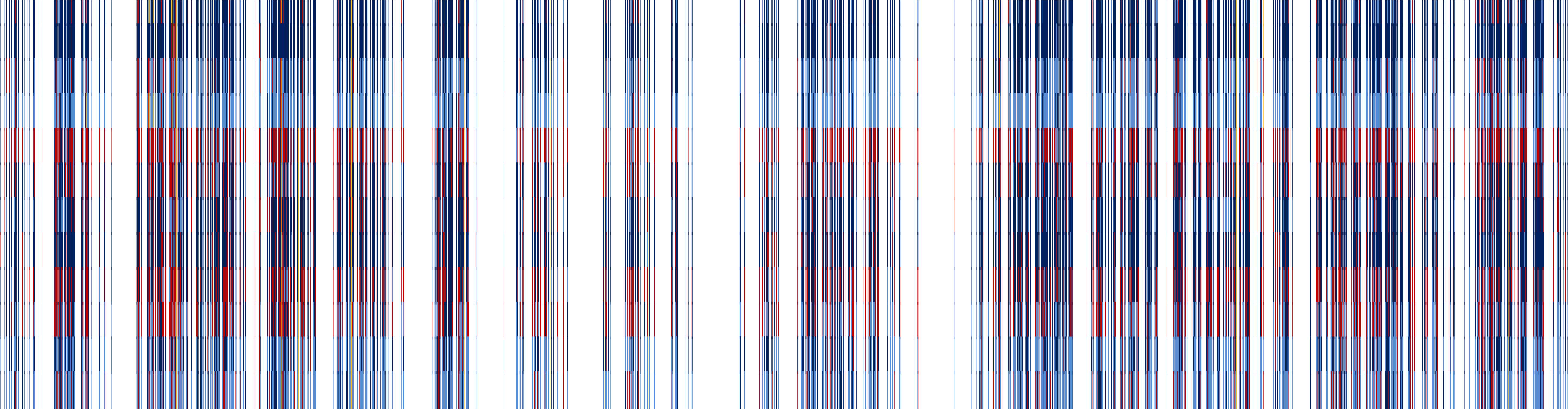
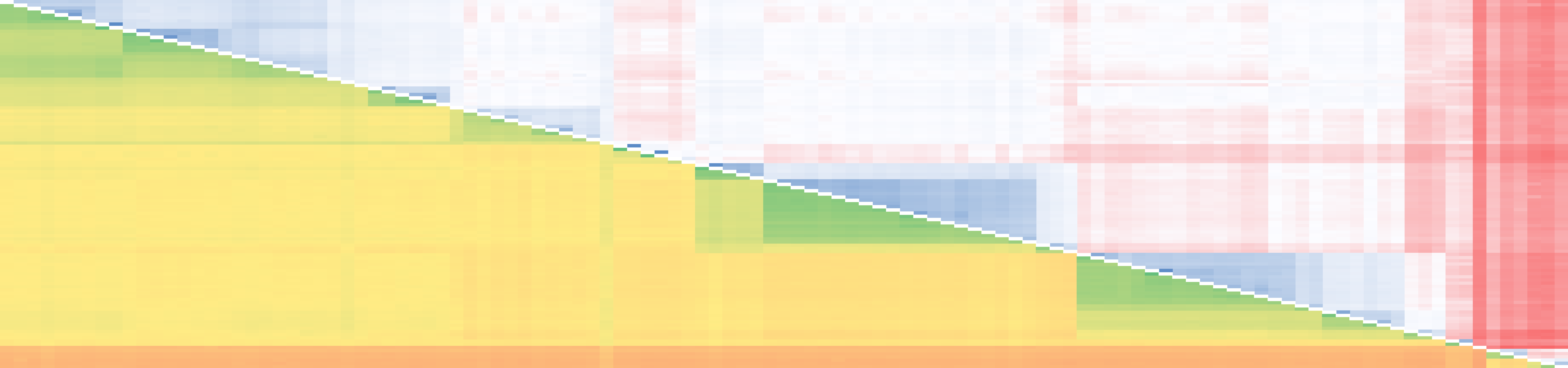
Microarray Screeing of pearl millet cDNA library. Note: Only 1/12 of Microarray image is shown. Green spots represent genes induced by pathogen elicitors. Orange/red spots represent genes repressed by elicitors.
Collaboration:
This project was initiated as the PhD project of Bridget Crampton, and has been supported by the African Centre for Gene Technologies. Click here for more information. Development of the SSH screening method was done in collaboration with Noelani van den Berg who applied the method to banana, and Dr. Paul Birch and Dr. Ingo Hein of the Scottish Crops Research Institute.
Crampton BG, Hein I, Berger DK (2009) Salicylic acid confers resistance to a biotrophic rust pathogen, Puccinia substriata in pearl millet, Pennisetum glaucum, Molecular Plant Pathology, 10 (2) 291-304
La w PJ, Claudel-Renard C, Joubert F, Louw AI, Berger DK (2008) MADIBA: A web server toolkit for biological interpretation of Plasmodium and plant gene clusters BMC Genomics 9: 105
Berger DK, Crampton B, Hein I, Vos W (2007). Screeing cDNA libraries on glass slide microarrays. In: Microarrays, Second Edition, Volume II Applications and Data Analysis (Editor J.Brampal), Series: Methods in Molecular Biology (Series Editor J.M.Walker), Humana Press, Totowa, New Jersey, USA (www.humanapress.com) ISBN10# 1-58829-944-9, Chapter 12, 177-203.
Van den Berg N, Crampton BC, Birch PRJ, Hein I & Berger DK (2004). High throughput screening of SSH cDNA libraries using DNA microarray analysis. Biotechniques. 37(5): 818-824 (November).