Pietersen G, Morgan S,
Read DA. (2024)
First report of Citrus concave gum-associated virus (CCGaV) on apple (Malus spp.) in South Africa.
Journal of Plant Pathology
10.1007/s42161-024-01629-9
Robert R, Robberste N, Thompson GD,
Read DA. (2024)
Characterization of macadamia ringspot‑associated virus, a novel Orthotospovirus associated with Macadamia integrifolia in South Africa.
European Journal of Plant Pathology
10.1007/s10658-024-02832-1 
Read DA, Thompson GD, Swanevelder DZH,
Pietersen G. (2023)
Metaviromic Characterization of Betaflexivirus Populations Associated with a Vitis cultivar Collection in South Africa.
Viruses 15(7):1474.
10.3390/v15071474
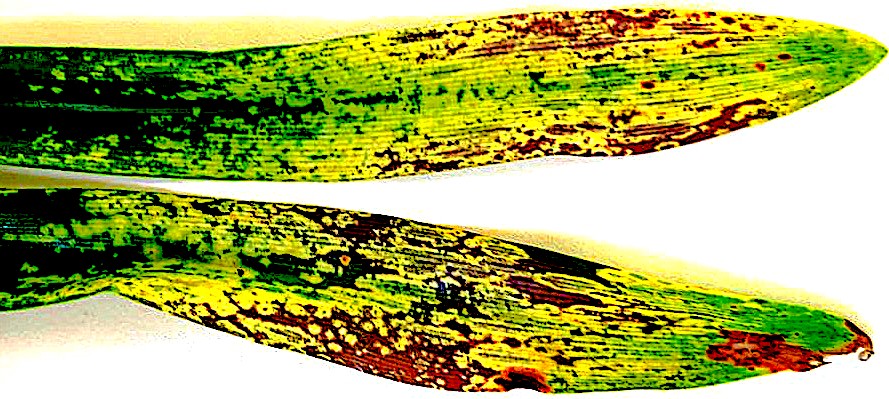
David Read, Ronel Roberts, Goddy Prinsloo, Genevieve Thompson. (2022)
Genomic and phylogenetic characterization of wheat yellows virus, a novel tenuivirus infecting wheat in South Africa.
Archives of Virology
10.1007/s00705-022-05649-7
Read DA, Thompson GD, Swanevelder D,
Pietersen G. (2021)
Detection and diversity of grapevine virus L from a Vitis cultivar collection in Stellenbosch, South Africa.
European Journal of Plant Pathology
10.1007/s10658-021-02380-y
Read DA,
Roberts R, Thompson GD. (2021)
Genomic characterization of two novel viruses infecting Barleria cristata L. from the genera Orthotospovirus and Polerovirus.
Archives of Virology
10.1007/s00705-021-05150-7
Kleynhans J,
Pietersen G. (2016)
Comparison of multiple viral population characterization methods on a candidate cross-protection Citrus tristeza virus (CTV) source.
Journal of Virological Methods 237:92-100.
10.1016/j.jviromet.2016.09.003
New SA, van Heerden SW,
Pietersen G, Esterhuizen LL. (2016)
First report of a Turnip Yellows virus in association with the Brassica Stunting Disorder in South Africa.
Plant Disease 100(11):2341.
10.1094/PDIS-12-15-1443-PDN
Lin H,
Pietersen G, Han C,
Read DA, Lou B, Gupta G, Civerolo EL. (2015)
Complete genome sequence of “Candidatus Liberibacter africanus,” a bacterium associated with Citrus Huanglongbing.
Genome Announcements 3(4):e00733-15.
10.1128/genomeA.00733-15 
Roberts R,
Steenkamp ET,
Pietersen G. (2015)
Three novel lineages of ‘Candidatus Liberibacter africanus’ associated with native rutaceous hosts of Trioza erytreae in South Africa.
International Journal of Systematic and Evolutionary Microbiology 65:723-731.
10.1099/ijs.0.069864-0
Zablocki O,
Pietersen G. (2014)
Characterization of a novel citrus tristeza virus genotype within three cross-protecting source GFMS12 sub-isolates in South Africa by means of Illumina sequencing.
Archives of Virology 159(8):2133-2139.
10.1007/s00705-014-2041-3
Almeida RPP, Daane KM, Bell VA, Blaisdell GK, Cooper ML, Herrbach E,
Pietersen G. (2013)
Ecology and management of grapevine leafroll disease..
Frontiers of Microbiology 4(94):1-13.
3389/fmicb.2013.00094 
Pietersen G, Spreeth N, Oosthuizen T, Van Rensburg A, Van Rensburg M, Lottering D, Rossouw N, Tooth D. (2013)
A case study of control of Grapevine Leafroll Disease spread at a commercial wine estate in South Africa.
American Journal of Enology and Viticulture 64:296-305.
10.5344/ajev.2013.12089 
Scott KA, Hlela Q,
Zablocki ODJ,
Read DA, Van Vuuren SP,
Pietersen G. (2013)
Genotype composition of populations of Citrus tristeza virus grapefruit cross-protecting GFMS12 in different host plants and aphid-transmitted sub-isolates.
Archives of Virology 158(1):2-37.
10.1007/s00705-012-1450-4 
Walsh HA,
Pietersen Gerhard. (2013)
Rapid detection of Grapevine leafroll-associated virus type 3 using a reverse transcription loop-mediated amplification method.
Journal of Virological Methods 194:308-316.

Phaladira MNB,
Viljoen R,
Pietersen G. (2012)
Widespread occurrence of “Candidatus Liberibacter africanus subspecies capensis” in Calodendrum capense in South Africa.
European Journal of Plant Pathology 124:39-47.

Rey MEC, Ndunguru J, Berrie LC, Paximadis A, Berry S, Cossa N, Nuaila VN, Mabasa KG, Abraham N, Rybicki EP, Martin DP,
Pietersen G, Esterhuizen L. (2012)
Diversity of Dicotyledenous-Infecting Geminiviruses and Their Associated DNA Molecules in Southern Africa, Including the South-West Indian Ocean Islands..
Viruses 4(9):1753-1791.
10.3390/v4091753 
Jooste AEC,
Pietersen G, Burger JT. (2011)
Distribution of grapevine leafroll associated virus-3 variants in South African vineyards.
European Journal of Plant Pathology 131(3):371-381.

Kruger K, De Klerk A, Douglas-Smit N, Joubert J,
Pietersen G, Stiller M. (2011)
Aster yellows phytoplasma in grapevines: identification of vectors in South Africa.
Bulletin of Insectology 64:S137-S138.

Jooste A, Maree H, Bellstedt D, Goszczynski D,
Pietersen G, Burger J. (2010)
Three genetic grapevine leafroll-associated virus 3 variants identified from South African vineyards show high variability in their 5'UTR.
Archives of Virology :1-10.

Lamprecht RL, Kasdorf GGF, Stiller M, Staples SM, Nel LH,
Pietersen G. (2010)
Soybean blotchy mosaic virus, a New Cytorhabdovirus Found in South Africa.
Plant Disease 94(11):1348-1354.
10.1094/pdis-09-09-0598 
Pietersen G, Arrebola E, Breytenbach JHJ, Korsten L, Le Roux HF, La Grange H, Lopes SA, Meyer JB, Pretorius MC, Schwerdtfeger M, Van Vuuren SP, Yamamoto P. (2010)
A Survey for .
Plant Disease 94(2):244-249.
10.1094/pdis-94-2-0244
De Graca JV,
Pietersen G, Van Vuuren SP. (2009)
Research on HLB in South Africa.
Phytopathology 99:S157.
Doddapaneni H, Liao H, Lin H, Bai X, Zhao X, Civerolo EL, Irey M, Coletta-Filho H,
Pietersen G. (2008)
Comparative phylogenomics and multi-gene cluster analyses of the Citrus Huanglongbing (HLB)-associated bacterium 'Candidatus Liberibacter'.
BMC Research Notes 1(75)

Lamprecht R,
Pietersen G, Kasdorf G, Nel L. (2008)
Characterisation of a proposed Nucleorhabdovirus new to South Africa.
European Journal of Plant Pathology 123(1):105-110.
10.1007/s10658-008-9339-5 
Meyer JB, Kasdorf GGF, Nel LH,
Pietersen G. (2008)
Transmission of activated-episomal Banana streak OL (badna)virus (BSOLV) to cv. Williams banana (Musa sp.) by three mealybug species.
Plant Disease 92(8):1158-1163.
Saccaggi DL, Ger K,
Pietersen G. (2007)
A multiplex PCR assay for the simultaneous identification of three mealybug species (Hemiptera: Pseudococcidae).
Bulletin of Entomological Research 98:27-33.

Stewart K,
Pietersen G. (2007)
Dynamics and molecular characterization of citrus tristeza virus (CTV) strains within the South African GFMS 12 cross-protecting population.
South African Journal of Science 103(1-2):II-II.
Pietersen G. (2004)
Spread of grapevine leafroll disease in South Africa-a difficult but not insurmountable problem.
Wineland (South Africa)
Pietersen G, Morris J. (2002)
Natural occurrence of groundnut ringspot virus on soybean in South Africa.
Plant Disease 86(11):1271.
Pietersen G, Smith MF. (2002)
Tomato yellow leaf curl virus resistant tomatoes show resistance to Tomato curly stunt virus.
Plant Disease 86(5):528-534.
Jooste AEC,
Pietersen G, Kasdorf GGF, Goszczynski DE. (2001)
Identification of three viruses from Pisum sativum in South Africa.
African Plant Protection 7(2):59-65.
Pietersen G, Idris AM, Kruger K, Brown JK. (2000)
Tomato curly stunt virus, a New Begomovirus of Tomato Within the Tomato yellow leaf curl virus-IS Cluster in South Africa.
Plant Disease 84(7):810-810.
10.1094/pdis.2000.84.7.810b
Cook G, Kasdorf GGF,
Pietersen G. (1999)
Synopsis of a virus survey on groundnut in South Africa.
African Plant Protection 5(1):1-3.