The advent of next-generation sequencing (NGS) technologies has revolutionised research capacity and its broad applicability over the past decade. These technologies have directly impacted researchers’ ability to understand host-pathogen interactions, holistically, and on a molecular scale. Recently, the avocado industry has begun to unravel some of the underlying mysteries that pertain to prevalent avocado pests and diseases; the Avocado Research Programme (ARP) has been highly influential in this space.
As a founding member of the Avocado Genome Consortium - an international collaborative effort which was established in 2016 - the ARP has directly contributed to the development of a high quality, chromosome-level reference genome assembly for Persea americana (avocado). The Avocado Genome Consortium has used this new reference genome to re-sequence more than 10 additional cultivars and rootstocks as part of an ongoing project, with plans to increase this number soon. Provided with this data, researchers will be able to accelerate the arduous process of selecting rootstocks and cultivars with desirable traits.
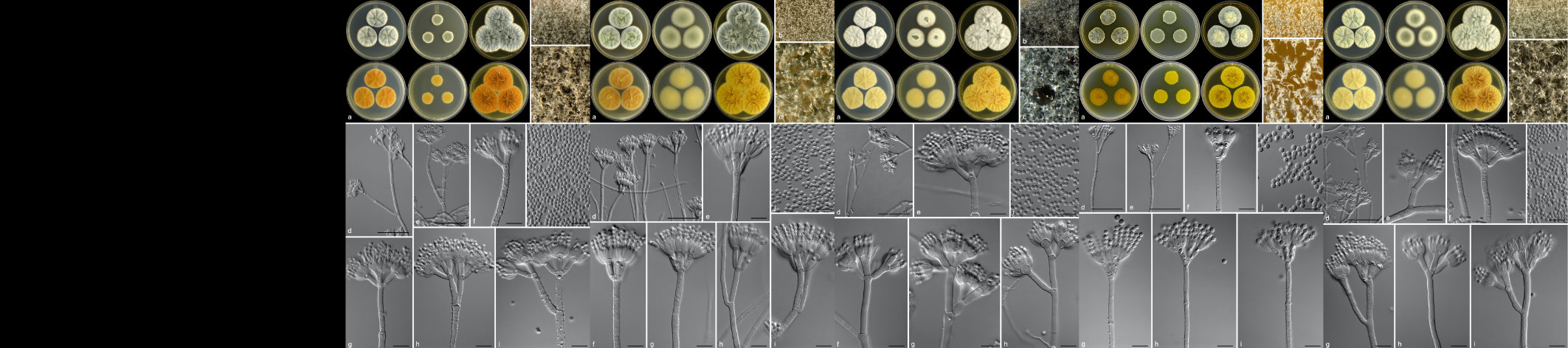
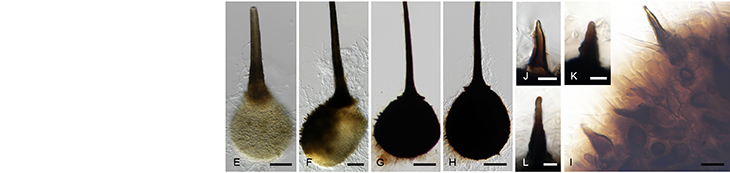
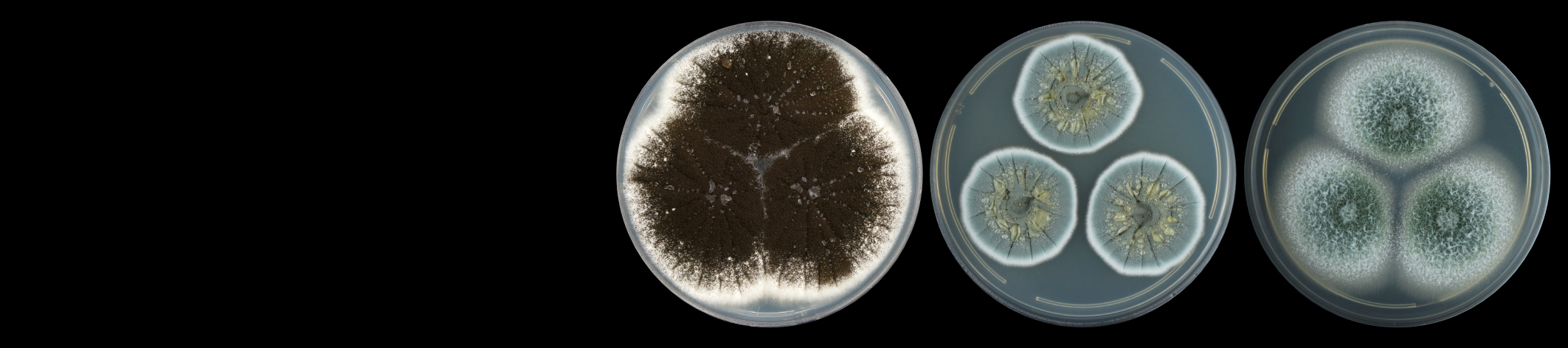
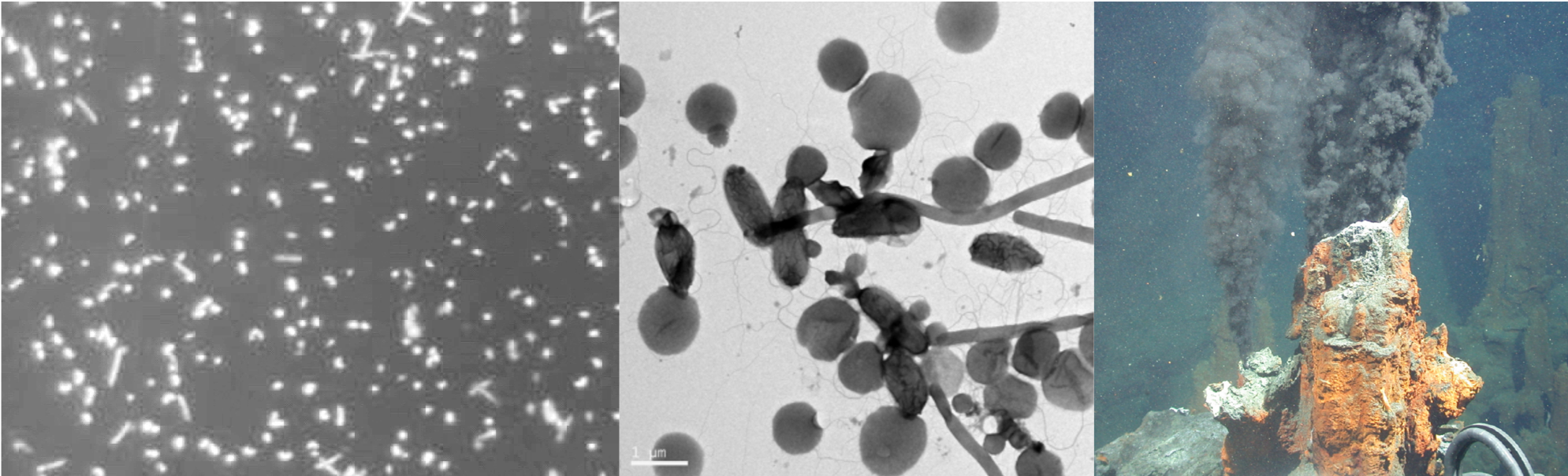
As part of the broader objectives of the Avocado Genome Consortium, transcriptomic data has also been generated. This data will be utilized to answer fundamental questions pertaining to the evolutionary biology, gene expression, physiological processes, and molecular pathways in avocado. Transcriptomic data from a dual RNA-sequencing experiment - involving both susceptible and partially resistant avocado rootstocks challenged with Phytophthora cinnamomi - was also used to identify avocado defence targets and discovery of pathogen effectors involved in disease development. The data from this work has been published across several research articles and will be used to further our understanding of the avocado-P. cinnamomi interaction.
The researchers involved are:
- David Kuhn, USDA, Florida
- Patricia Manosalva, UCR, California
- Noëlani van den Berg, UP, South Africa
- Antonio Javier Matas Arroyo, Departamento de Biología Vegetal, University of Malaga, Spain
- Aureliano Bombarley Gomez, Virginia Tech Horticulture, USA
- Randy Ploetz, University of Florida, USA
- Alan Chambers, University of Florida, USA