Eucalyptus synthetic biology resources for re-engineering woody biomass for biomaterials and bioenergy
The ability to assemble standardized biological (DNA) parts idempotently is a core feature of synthetic biology. Recently, the Phytobrick standard (Patron et al., 2015) incorporating a universal syntax enabling one-step assembly of level 0 eukaryotic gene elements (i.e. promoters, 5' UTRs, coding sequences, etc) into transcriptional units has gained acceptance in the plant synthetic biology community. These standards are compatible with modular cloning (MoClo), Golden Gate, GoldenBraid, Gibson and LOOP DNA assembly methods (Engler et al. 2008, 2014; Gibson et al. 2009; Pollak et al. 2018; Sarrion-Perdigones et al. 2011, 2013; Weber et al. 2011), allowing scarless, versatile, hierarchical and one-pot assembly of large multigene constructs.
The US Department of Energy Joint Genome Institute (DOE-JGI), through a DNA Synthesis Science Program grant (CSP-503083), has funded the synthesis of 221 secondary cell wall-related E. grandis transcription factors and 65 promoter sequences to enable bioenergy and biomaterials-focused research. Specifically, we designed the synthetic panel for high-throughput protein-DNA interaction screening (e.g. DAP-seq; eY1H) (Bartlett et al., 2017; Gaudinier et al., 2011), in addition to functional genomics applications. Moreover, the majority of the synthetic constructs were designed as standardized Phytobricks, with additional GATEWAY functionality (Fig. 1; Hussey et al., in preparation). The full list of DNA constructs can be downloaded here.
The University of Pretoria (as Recipient) has a Materials Transfer Agreement (MTA) in place with the Regents of the University of California through the DOE-JGI (Contractor) regarding the use and transfer of constructs to third parties. The materials are freely available to nonprofit universities and institutions, subject to an MTA between that institution and the University of Pretoria. For any queries regarding the constructs or requests for material transfer, please contact This email address is being protected from spambots. You need JavaScript enabled to view it..
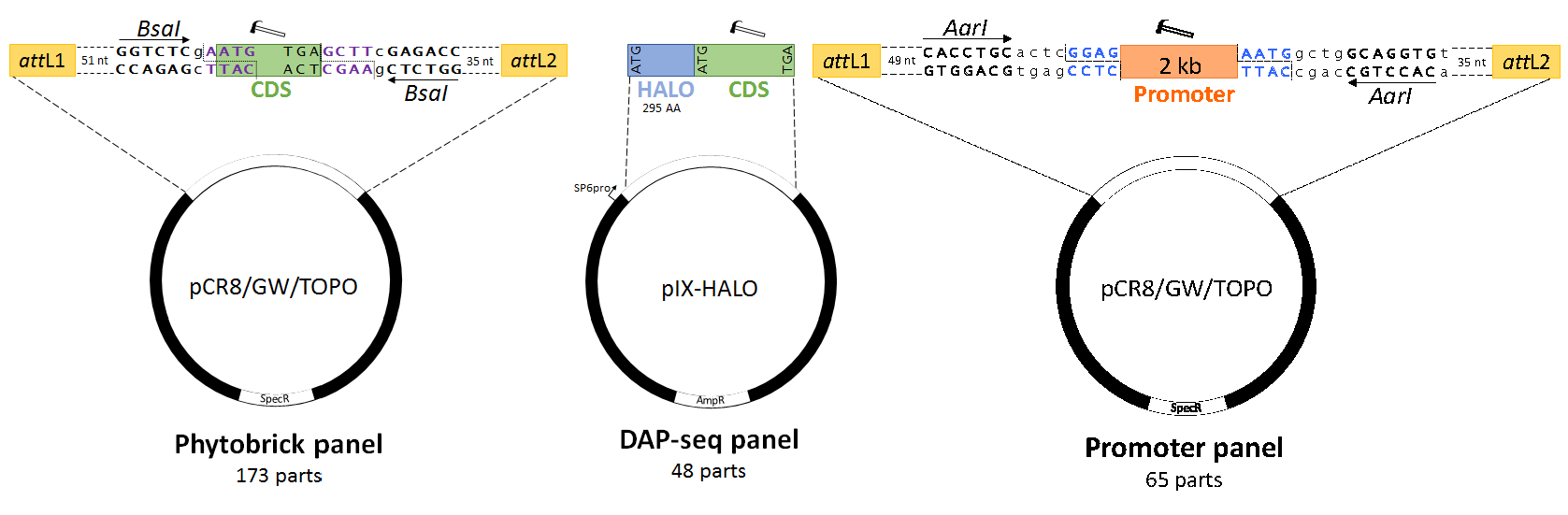
Figure 1. Design of standardized synthetic E. grandis SCW-related transcription factors and promoters. Hammers indicate domesticated sequences, dotted lines indicate restriction cleavage sites, coloured bases indicate standard syntax sequences and lowercase sequences indicate spacer nucleotides. (A) Transcription factor Phytobricks contain attL GATEWAY recombination sites (green), fifty-one basepair chewback linkers, BsaI Type IIS recognition sites and standard syntax sequences (purple text). The start codon of the domesticated coding sequence (green) remains in frame with N-terminal tags in GATEWAY destination vectors, while being primarily intended for Golden Gate, MoClo and Golden Braid assembly. (B) The DNA Affinity Purification Sequencing (DAP-seq) panel of transcription factors is available as a C-terminal fusion to the HALO purification tag, intended for in vitro transcription and translation via the SP6 phage promoter. While not standardized by a universal syntax, the coding sequences are fully domesticated and can thus be subcloned as standardized parts into a universal acceptor plasmid. (C) The secondary cell wall promoter panel features semi-domesticated 2 kb promoter sequences (including 5’ UTRs) compatible with GATEWAY and AarI-mediated Golden Gate cloning. Standard prefix and suffix syntax sequences allow for two-step Type IIS assembly to any Phytobrick panel CDS in (A). AmpR, ampicillin resistance gene; CDS, coding sequence; SpecR, spectinomycin resistance gene.
References
Bartlett, A., O'Malley, R.C., Huang, S.C., Galli, M., Nery, J.R., Gallavotti, A. and Ecker, J.R. (2017) Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat Protoc 12, 1659-1672.
Engler, C., Kandzia, R. and Marillonnet, S. (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS One 3, e3647.
Engler, C., Youles, M., Gruetzner, R., Ehnert, T.M., Werner, S., Jones, J.D., Patron, N.J. and Marillonnet, S. (2014) A golden gate modular cloning toolbox for plants. ACS Synth Biol 3, 839-843.
Gaudinier, A., Zhang, L., Reece-Hoyes, J.S., Taylor-Teeples, M., Pu, L., Liu, Z., Breton, G., Pruneda-Paz, J.L., Kim, D., Kay, S.A., Walhout, A.J., Ware, D. and Brady, S.M. (2011) Enhanced Y1H assays for Arabidopsis. Nat Methods 8, 1053-1055.
Gibson, D.G., Young, L., Chuang, R.Y., Venter, J.C., Hutchison, C.A., 3rd and Smith, H.O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343-345.
Patron, N.J., Orzaez, D., Marillonnet, S., Warzecha, H., Matthewman, C., Youles, M., Raitskin, O., Leveau, A., Farré, G., Rogers, C., Smith, A., Hibberd, J., Webb, A.A., Locke, J., Schornack, S., Ajioka, J., Baulcombe, D.C., Zipfel, C., Kamoun, S., Jones, J.D., Kuhn, H., Robatzek, S., Van Esse, H.P., Sanders, D., Oldroyd, G., Martin, C., Field, R., O'Connor, S., Fox, S., Wulff, B., Miller, B., Breakspear, A., Radhakrishnan, G., Delaux, P.M., Loqué, D., Granell, A., Tissier, A., Shih, P., Brutnell, T.P., Quick, W.P., Rischer, H., Fraser, P.D., Aharoni, A., Raines, C., South, P.F., Ané, J.M., Hamberger, B.R., Langdale, J., Stougaard, J., Bouwmeester, H., Udvardi, M., Murray, J.A., Ntoukakis, V., Schäfer, P., Denby, K., Edwards, K.J., Osbourn, A. and Haseloff, J. (2015) Standards for plant synthetic biology: a common syntax for exchange of DNA parts. New Phytol 208, 13-19.
Pollak, B., Cerda, A., Delmans, M., Álamos, S., Moyano, T., West, A., Gutiérrez, R.A., Patron, N., Federici, F. and Haseloff, J. (2018) Loop Assembly: a simple and open system for recursive fabrication of DNA circuits. bioRxiv, 247593.
Sarrion-Perdigones, A., Falconi, E.E., Zandalinas, S.I., Juarez, P., Fernandez-del-Carmen, A., Granell, A. and Orzaez, D. (2011) GoldenBraid: an iterative cloning system for standardized assembly of reusable genetic modules. PLoS One 6, e21622.
Sarrion-Perdigones, A., Vazquez-Vilar, M., Palaci, J., Castelijns, B., Forment, J., Ziarsolo, P., Blanca, J., Granell, A. and Orzaez, D. (2013) GoldenBraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol 162, 1618-1631.
Weber, E., Engler, C., Gruetzner, R., Werner, S. and Marillonnet, S. (2011) A modular cloning system for standardized assembly of multigene constructs. PLoS One 6, e16765.

