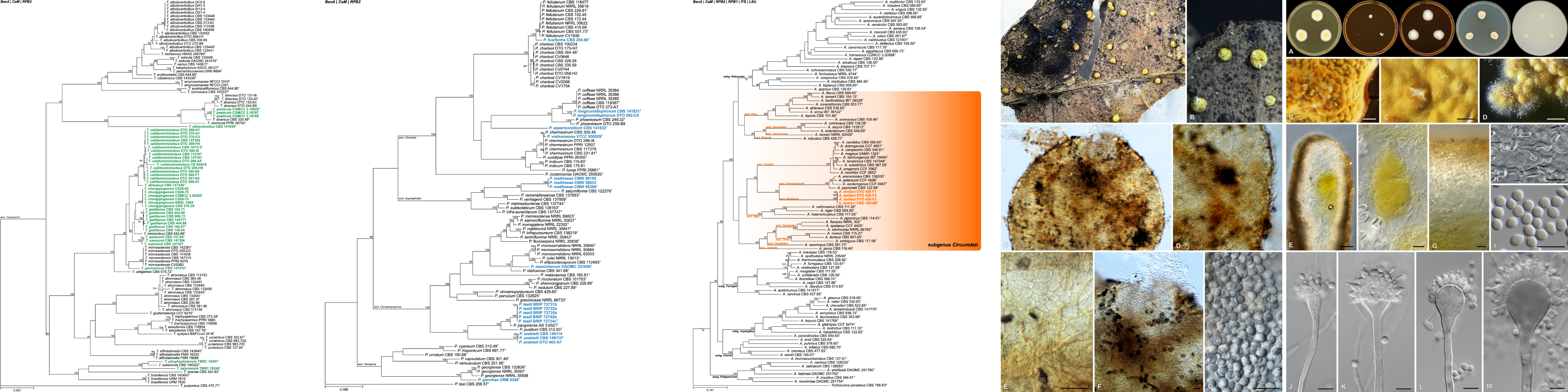
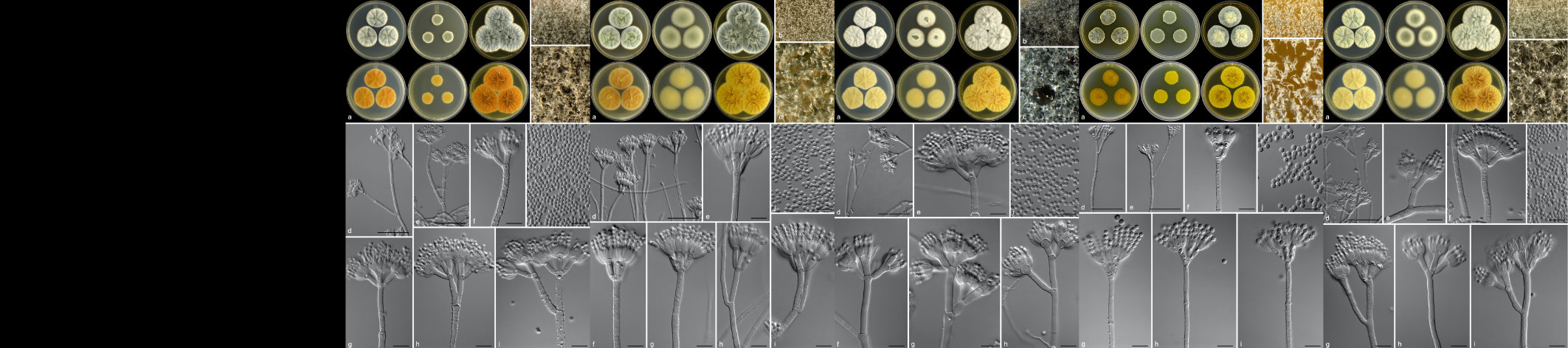
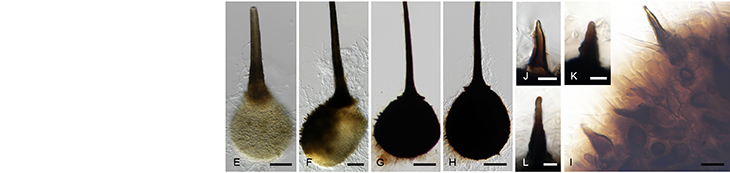
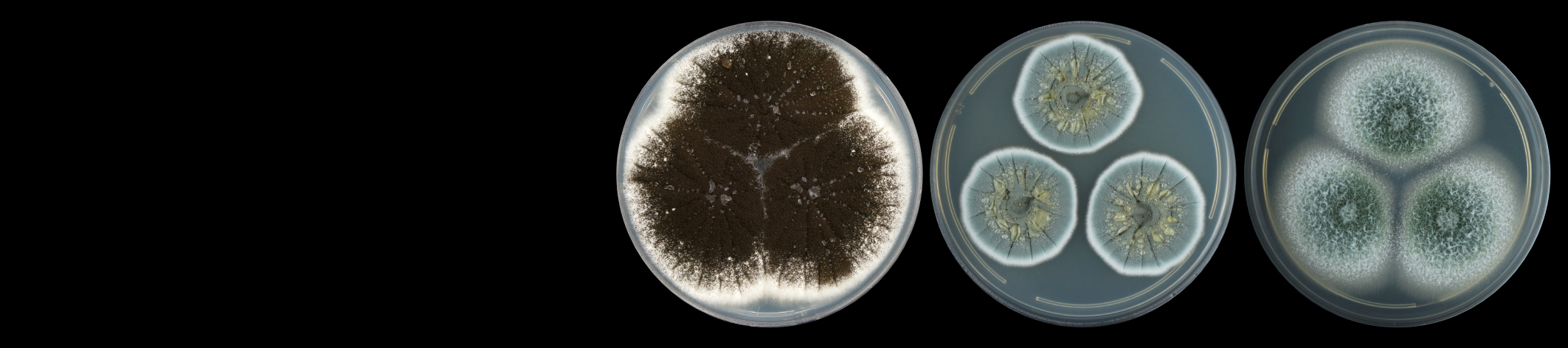
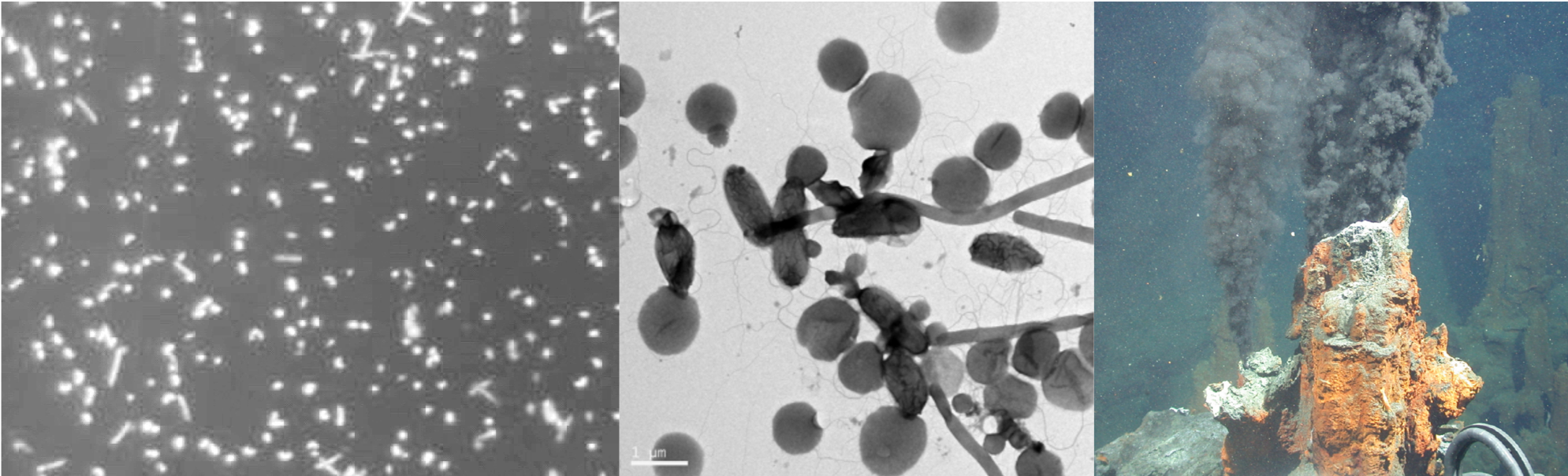
The FMG Bioinformatics Platform is hosted in the Centre for Bioinformatics and Computational Biology at the University of Pretoria. Our team consists of a MIT big data science MSc student, Mr Christopher Marais, and a postdoctoral fellow, Dr Nanette Christie.
Working alongside biologists and geneticists in the FMG Programme, we are able to support and build capacity for the analysis, management and interpretation of large amounts of genomics and transcriptomics data for hundreds to thousands of individual trees. Web services, novel and commercial software, as well as a Galaxy workflow environment have been implemented to facilitate modular development of data processing and visualisation pipelines. These pipelines can be accessed remotely by group members for high-throughput genomics analysis and enable quality control, processing of raw data and downstream analyses such as gene and genome annotation, gene expression analysis, detection of genomic variants, and discovery of genetics associations with phenotypic traits.
We worked with colleagues at the Umea Plant Science Centre in Sweden on the integration of the Eucalyptus Genome Integrative Explorer (EucGenIE) into PlantGenIE (plantgenie.org), a dedicated web portal for genome and transcriptome exploration in plants (also including poplar, Norway spruce, and Arabidopsis). We also developed a web resource to query and visualise Eucalyptus systems genetics data, such as expression quantitative trait locus (eQTL) networks. We envision our genetic variation tool (qtlXplorer) to be a significant building block towards a population genomics module in PlantGenIE.
We are also hosting a genomic breeding database (BioPlasm) providing data access and analysis tools for tree breeders of our industry partners.