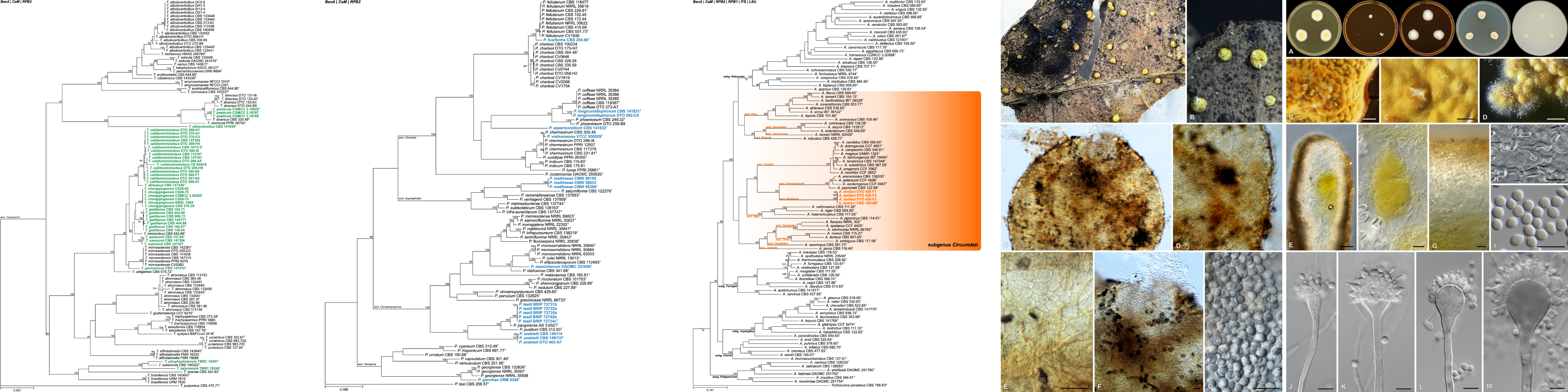
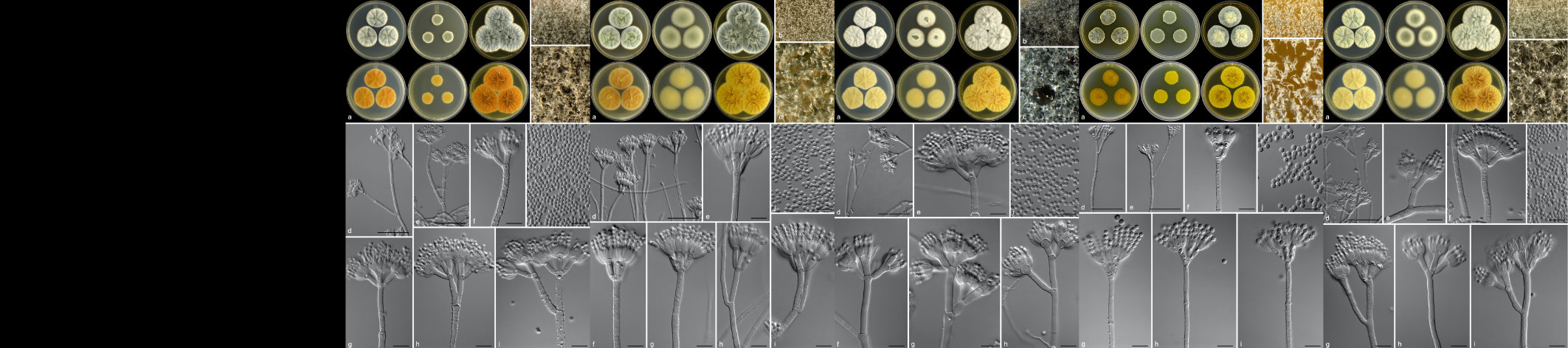
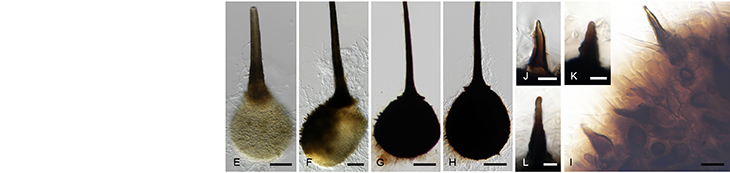
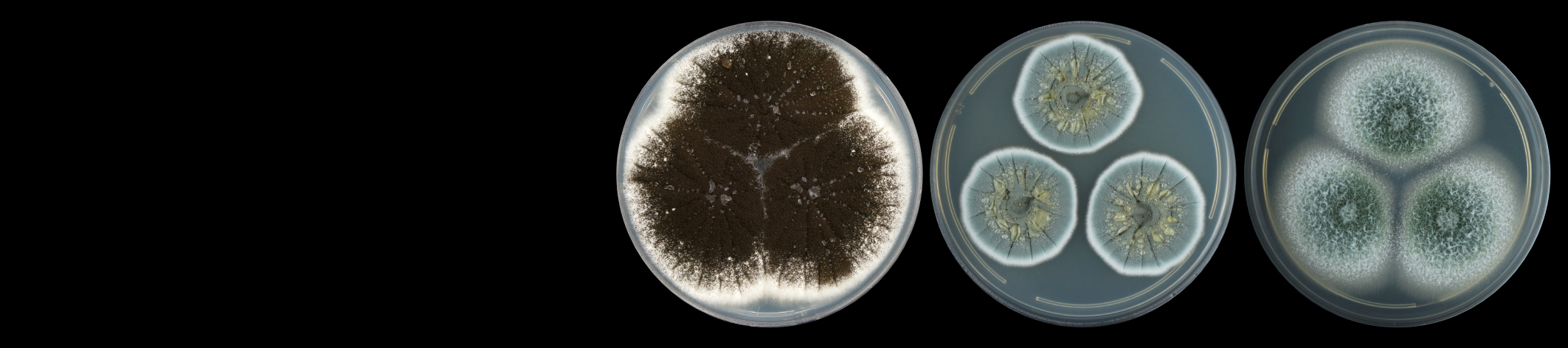
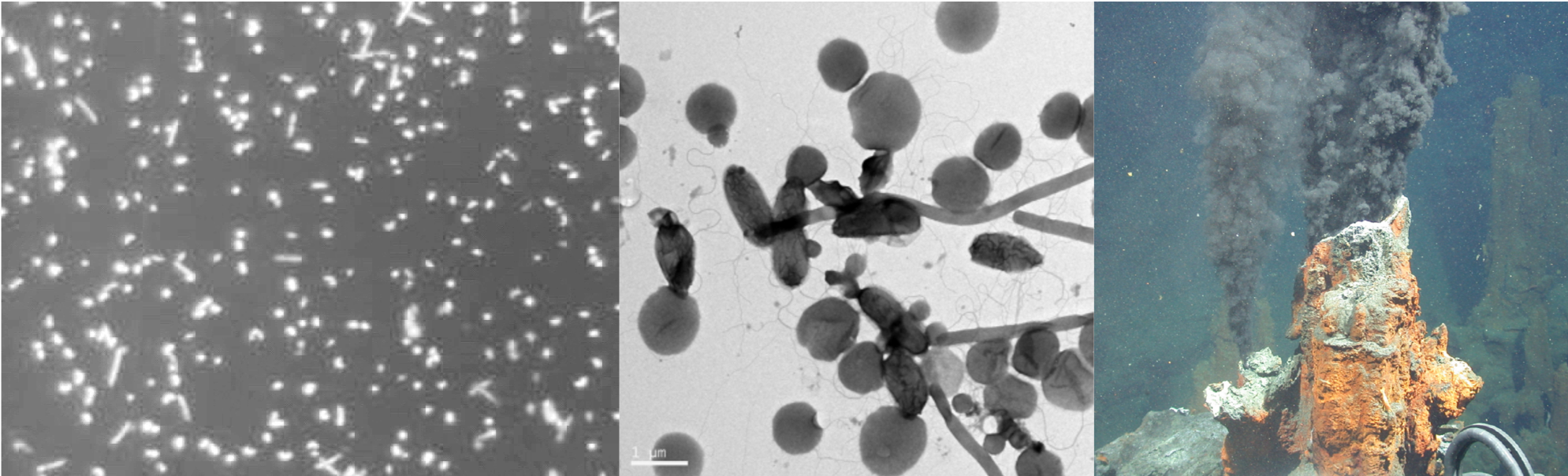
Quantitative Real-Time PCR is an extensively used analytical method in molecular biology research. With a grant from the NRF National Equipment
Platform (NEP) the QuantStudio™12K Flex Real-Time PCR Facility was established in 2014, aimed towards the provision of a user-friendly real-time PCR
platform to researchers throughout South Africa and to promote realtime PCR-based research. The QuantStudio System allows for small to large-scale
functional genomics studies, with two main functions, gene expression analysis or genotyping assays, being used in either 384 well format or high
throughput OpenArray® technology. As such the QuantStudio™12K Flex Real-Time PCR Facility has become an indispensable tool used by
staff and students in FMG, the university and surrounding universities across South Africa.
Specifically in FMG, functional genomic characterization of wood development and disease resistance projects benefit from the facility. These projects
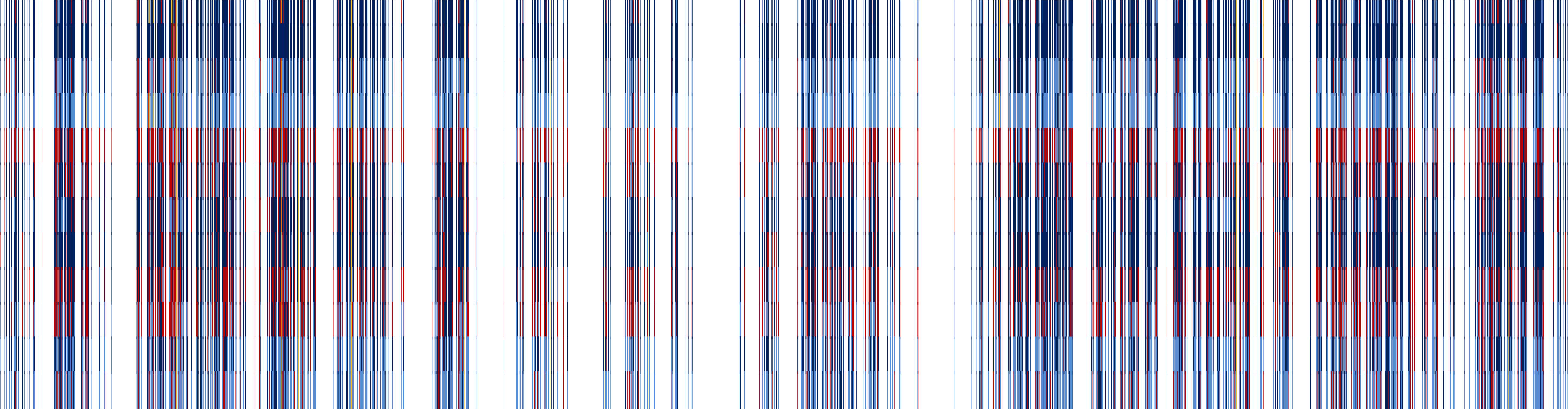
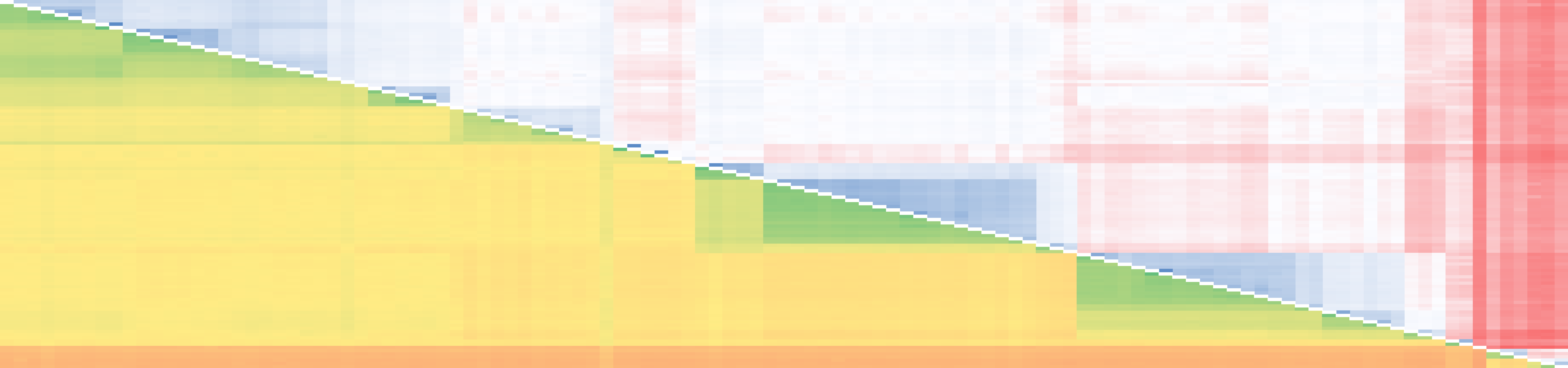
include the expression analysis of hundreds of genes under varying conditions and treatments, as well as SNP marker genotyping of up to 256 SNP markers
across thousands of individuals. In the past, FMG as a group has used custom designed assays for SNP marker genotyping, and analysis of gene expression
of pathogen defence related genes in Eucalyptus trees. Currently FMG uses the platform to investigate genes involved in secondary cell wall development in
Populus, understanding gene expression of nuclear vs plastid genomes, and several 384 well studies looking at genes of interest. Since the establishment
of the facility the QuantStudio™12K Flex Platform has encouraged collaboration and mentorship across a wide range of research groups, universities
and institutes and has been used by researchers in many different fields whether they are studying the metabolism of humans or researching drought
tolerance in cassava.