Principal Investigator: Prof Dave Berger
Current team members at UP: Dr Boney Kuriakose, Dr Sonia Phillips , Dr Maryke Carstens, Jeanne Korsman, Nanette Coetzer, Adri Veale, Grieta Mahlangu, Monique Heystek, Thomas Schmidt, Ncobile Kunene, Irene Schoeman.
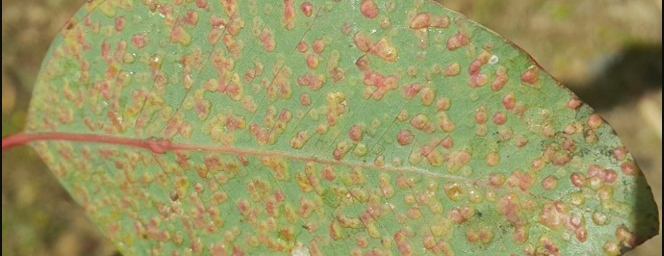
The project aims to identify genes and molecular markers linked to quantitative resistance to grey leaf spot disease in African maize varieties. The innovation is to combine classical quantitative genetic analysis, genetic mapping and global gene expression profiling. The power of including expression profiling using oligonucleotide microarrays is that it facilitates the identification of genes contributing to QTL for GLS resistance. Candidate genes and/or tightly linked molecular markers will be of enormous value to maize breeders for use in marker assisted selection of resistance traits. This project combines the skills of a number of experts in South Africa, and is one of the first expression QTL projects in the country. Establishing capacity in this technique will enable future work on additional favourable traits in crops for which global gene expression data are available.
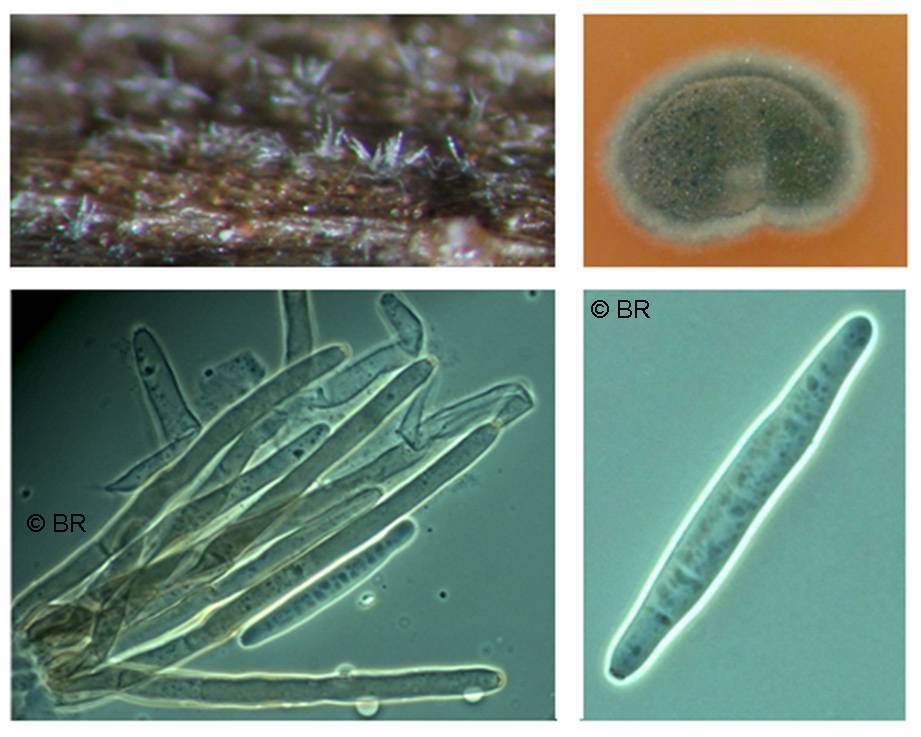
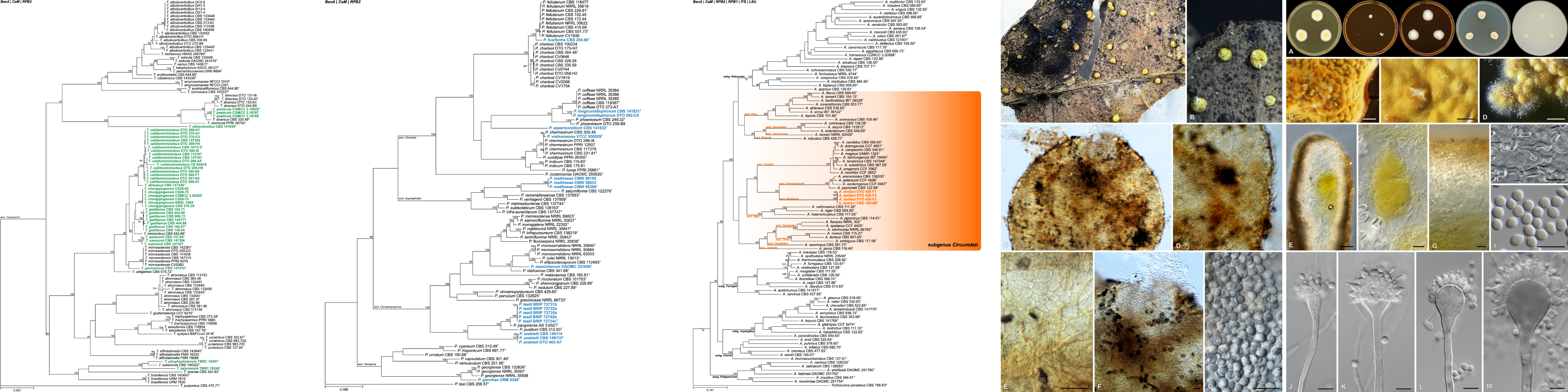
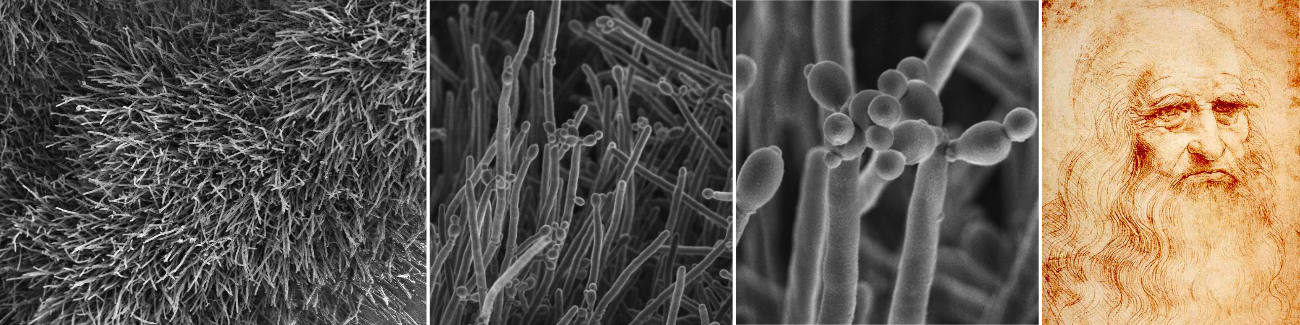
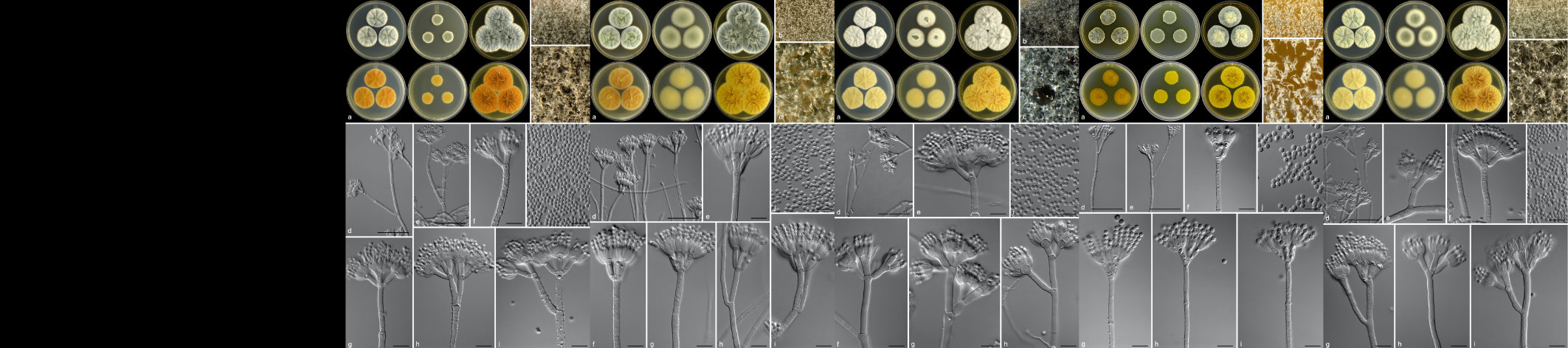
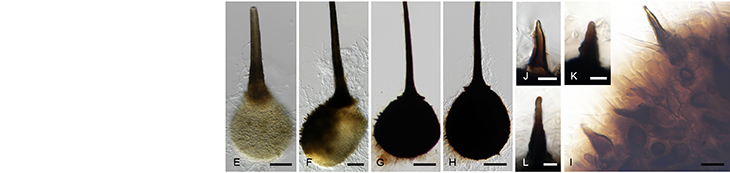
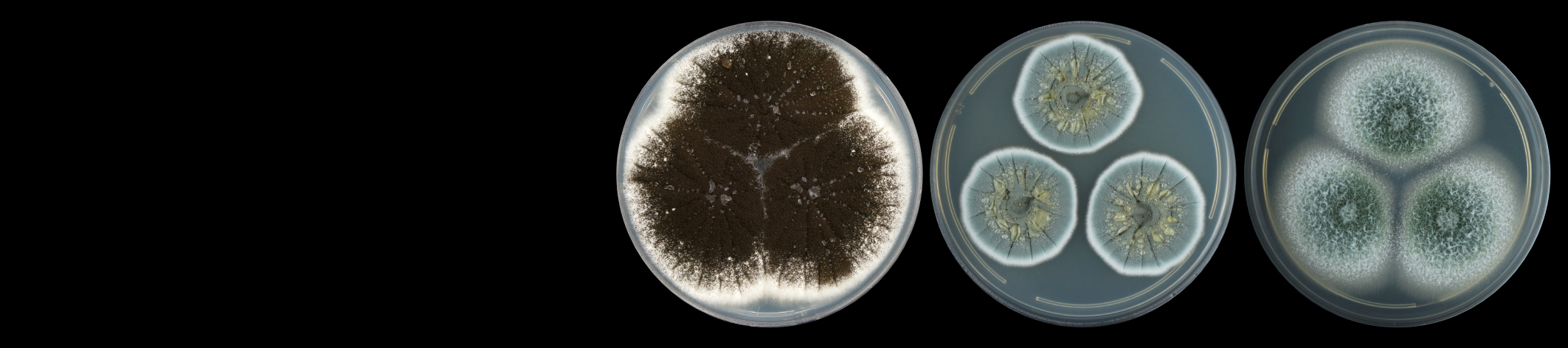
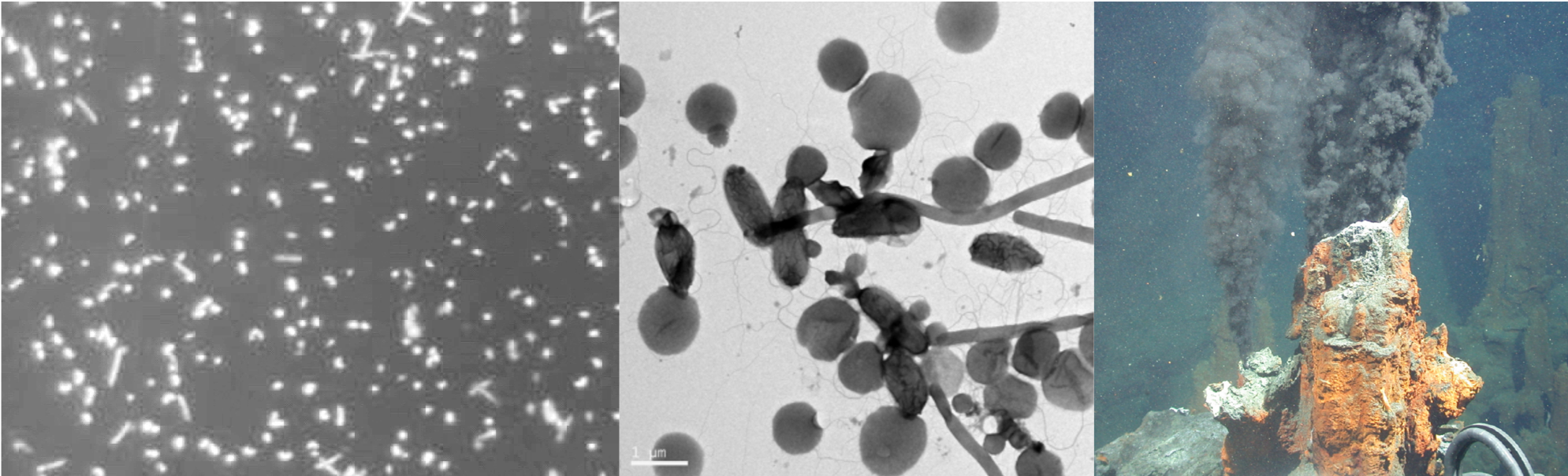
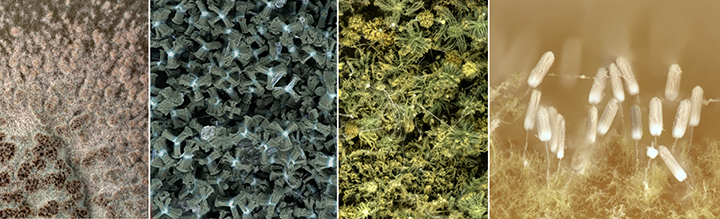
Cercospora zeina cultures with emphasis on the conidiophores and conidia
Maize eQTL project team

Collaborators
- African Centre for Crop Improvement (University of Kwa-Zulu Natal)
- Centre for Proteomic and Genomic Research (University of Cape Town)
- PANNAR
MeQTL project field trip

Standing L - R: Felix Middelton (PANNAR), Monique Heystek (UP), Thomas Schmidt (UP), Mischa Muller (UP), Dave Berger (UP), Hlandiwe Gumede (UKZN ACCI)
Seated L - R: Shane Murray (CPGR), Bridget Crampton (UP)
Publications:
-
Korsman J, Meisel B, Kloppers FJ, Crampton BG and Berger DK (2012) Quantitative phenotyping of grey leaf spot disease in maize using real-time PCR. European Journal of Plant Pathology, 133(2) 461-471
-
Coetzer N, Myburg AA & D K Berger (2011) Maize Microarray Annotation Database. Plant Methods 7:31.
-
Meisel B, Korsman J, Kloppers FJ and Berger DK (2009) Cercospora zeina is the causal agent of grey leaf spot disease of maize in southern Africa. European Journal of Plant Pathology, 124 (4) 577-583.